Global-to-local protein shape similarity system driven by digital elevation models
The article describing our global-to-local shape-similarity search system prototype has been published in the IEEE Biosmart 2017 conference
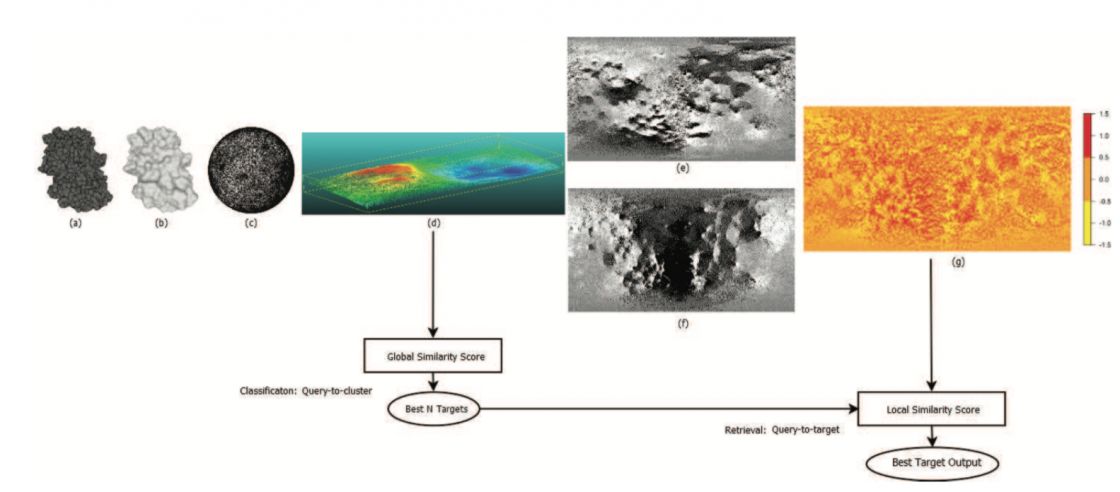
We are currently developing a bio-shape similarity system for supplying high-throughput protein shape similarity applications within massive datasets. The proposed system is powered by a global-to-local shape similarity system which exploits shape elevation and local convexity attributes. In the first step, a global similarity is computed between the shape descriptors associated to each protein input. The procedure outputs best N similarities chosen by the user, within a query-to-cluster approach. The second stage is a patch-based local similarity computation method which is designed to find the best similar target from the cluster for supplying query-to-target protein retrieval applications. The local patch-based similarity comparison benefits of a multi-CPU implementation, offering thus fast query search capabilities within massive datasets. Experimental results on the SHREC 2017 BioShape dataset composed of 5484 models, illustrate the effectiveness of the proposed system.
Dr. Daniela Craciun has presented the paper during the IEEE Biosmart conference on september 2017 in Paris, France
The article is available on the OpenAIRE section of the Zenodo Platform
